A platform technology in development for imaging enables the integration of microscopic analysis, long employed in pathology laboratories, with the visualization of multiple molecular markers in individual cells.
The research, published Thursday in Nature Cancer, indicates that such multiplex imaging may change cancer diagnostic testing by exposing molecular traits associated with a cancer’s aggressiveness or vulnerability to therapy.
Multiplex imaging has remained largely confined to academic labs because its methods are generally incompatible with the standardized tissue processing and staining involved in hematoxylin and eosin (H&E) microscopy, long employed in clinical pathology. Its methods also do not generally permit the evaluation of whole slides, a requirement of clinical diagnostics.
Clinical pathology relies on the microscopic analysis of tissues embedded in wax, sliced into strips about one-fifteenth the width of a human hair and stained pink and purple with H&E to expose the fine morphology of cells. Increasingly, researchers are applying machine learning and artificial intelligence algorithms to H&E-stained tissue images. Such work — enriched by the ability to draw from millions of archived H&E images — identifies patterns not readily apparent to the human eye, which can improve cancer diagnosis and management.
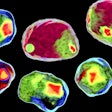
Meanwhile, the use of antibodies to capture the molecular characteristics of tumors has rapidly evolved and is now indispensable to cancer diagnostics. Researchers have recently begun simultaneously applying scores of antibodies to tissues, linking the immunofluorescence-based images generated this way to gene expression in cells, resulting in richly textured tumor portraits. With this molecular information, researchers can identify distinct cell types, determine their current state, and assess how they may be interacting with neighboring cells to promote tumor growth.
The researchers, working at Harvard Medical School and Seattle-based RareCyte, sought to develop a platform, named Orion, to bridge these parallel processes and to validate its use by both human experts and artificial intelligence algorithms in identifying molecular tumor features that may predict progression-free survival in colon cancer patients.
To test the platform, they analyzed colorectal cancer specimens from 40 patients to find molecular features most closely associated with poor outcomes. Sifting through about 15,000 biomarker combinations, they identified those most tightly linked to patient prognosis and applied them to samples from 34 other colorectal cancer patients with known outcomes. The researchers show that the selected biomarkers predicted with high accuracy — 19 times out of 20 — the likelihood of a poor prognosis.
Combining multiplex and H&E imaging revealed illuminating relationships between molecular markers, cell morphology, and tumor topography. One such finding indicated that inflammation or immune activity at the rim of the tumor is of pathological significance. Another revealed the molecular basis of cell adhesion — a tissue morphology associated with propensity for metastasis.
The researchers plan to continue establishing a proof of concept for their platform, exploring markers for factors including treatment response and resistance. Ultimately, they hope the discoveries made using Orion are validated in large clinical trials.
“The prospect of incorporating high-powered advanced imaging tools to analyze routine clinical samples is very exciting,” Sandro Santagata, associate pathology professor at Brigham and Women’s Hospital, Harvard Medical School, said in a statement. “It promises to unlock a wealth of knowledge about cancer tissues.”