Seegene Technologies' Novaplex SARS-CoV-2 polymerase chain reaction (PCR) assays can quickly and reliably detect SARS-CoV-2 in patient samples and identify known variants with an accuracy comparable to the results from the "gold standard" Sanger sequencing method of the virus's spike gene, according to a recent study published in the Journal of Molecular Diagnostics.
The Novaplex variant I, II, and IV PCR assays are designed to detect genetic mutations associated with the alpha, beta, delta, and epsilon variants of SARS-CoV-2 (at the time of the study, the omicron variant had not yet emerged).
The researchers, led by Marisa Nielsen, PhD, of the department of pathology at the University of Texas Medical Branch in Galveston, said that the assays are accurate and less labor-intensive.
"The I, II, and IV assays provide an accurate, rapid, and less labor-intensive method for detecting SARS-CoV-2 and identifying known variants of interest and concern," they said.
RNA extraction vs. extraction-free protocols
The study evaluated both RNA extraction and extraction-free versions of the assays. The extraction-free protocols, in which samples are processed without extracting RNA for testing, are attractive because they significantly reduce the cost and turnaround time of COVID testing, the authors noted. In the study, the extraction-free protocol was able to generate results at least one day sooner than the standard chloroform-ethanol RNA extraction protocol (J Mol Diagn, February 23, 2022).
"RT-PCR [reverse transcription-PCR] methodology for variant detection is accessible, rapid, simpler, and accurate compared to traditional sequencing," said lead investigator Ping Ren, PhD, of the department of pathology at the University of Texas Medical Branch, in a statement. "Combining an extraction-free processing method with RT-PCR technology can help laboratories without sequencing capabilities track circulating variants and investigate variant-dependent effects on treatment efficacy and disease severity."
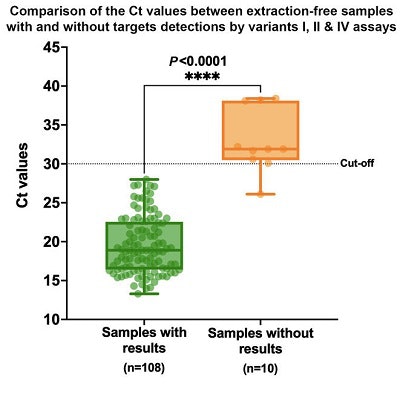
When the RNA extraction protocol was used, there was 100% overall agreement between the spike gene sequencing and the Novaplex results for the variants included in the Novaplex assay menus. The RNA extraction-free method was 91.7% as sensitive as the traditional RNA extraction method but produced results much more quickly and required no reagents.
 A higher cycle threshold (Ct) value indicates a lower viral load in the samples with and without results by variants I, II, and IV assays using the extraction-free method. Some mutations were detected less frequently in samples with lower viral load. Image courtesy of Seegene.
A higher cycle threshold (Ct) value indicates a lower viral load in the samples with and without results by variants I, II, and IV assays using the extraction-free method. Some mutations were detected less frequently in samples with lower viral load. Image courtesy of Seegene."Although lower sensitivity was observed with the extraction-free method, it still represents a viable alternative," said Nielsen. "A major limiting factor for molecular SARS-CoV-2 assays is the shortage of RNA extraction reagents, and conventional extraction remains a time-consuming aspect of molecular diagnosis of SARS-CoV-2."
After the study was completed, Seegene released the Novaplex SARS-CoV-2 Variants VII assay, which identifies the omicron mutation.
In addition, the authors noted that spike sequencing is still necessary for detecting new variants.
As new variants emerge, RT-PCR assays can be tailored to include additional representative genes. Rapid detection and monitoring of specific variants are necessary to inform public health and treatment decisions, according to the authors.
"Determining the SARS-CoV-2 variant in individual patient samples can help guide treatment since some variants are more resistant to current treatment regimens," Ren said. "However, the potential impact extends beyond individual patients and into the public health realm. It is important to track variant spread as part of public health surveillance because of variant-dependent transmission, disease severity, and treatment decisions."