A deep-learning system developed at Google outperformed general pathologists for Gleason grading of prostate cancer biopsies, company researchers reported in JAMA Oncology online July 23.
The research comes from a deep-learning/artificial intelligence (AI) research team at Google Health in Palo Alto, CA. For the study, performance of the deep-learning system and general pathologists were compared to assessment by a panel of urologic subspecialist pathologists for the evaluation of prostate cancer needle core biopsy formalin-fixed paraffin-embedded (FFPE) specimens.
In a validation set of 498 specimens that were positive for cancer, the deep-learning system came up with the same result as the panel of sub-specialists in 71.7% of cases, versus 58% for general pathologists, a statistically significant result.
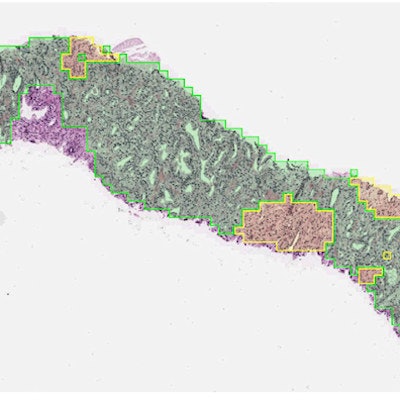
 On digitized images of prostate biopsies, the deep-learning system assigns Gleason patterns to every region (e.g., pattern 3 in green and pattern 4 in yellow in the figure). Next, the system summarizes these observations as a slide-level Gleason score and corresponding grade group. Image courtesy of Yun Liu, PhD, Google Health.
On digitized images of prostate biopsies, the deep-learning system assigns Gleason patterns to every region (e.g., pattern 3 in green and pattern 4 in yellow in the figure). Next, the system summarizes these observations as a slide-level Gleason score and corresponding grade group. Image courtesy of Yun Liu, PhD, Google Health."The [deep-learning system] warrants evaluation as an assistive tool for improving prostate cancer diagnosis and treatment decisions, especially where subspecialist expertise is unavailable," wrote Dr. Craig Mermel, PhD, product lead for pathology at Google Brain, and colleagues.
Gleason grading an 'imperfect diagnostic tool'
Deep learning has potential to play a role in an area of pathologic evaluation of prostate biopsies that needs improvement -- Gleason grading, which is an imperfect diagnostic tool, the authors wrote. Grading of prostate biopsy specimens to indicate clinical risk is based on subjective assessment of various patterns, such as pattern 5, which represents poorly differentiated cells, they noted.
"Consequently, it is common for different pathologists to assign a different [Gleason grade group] to the same biopsy (30%- 50% discordances)," Mermel et al wrote. "In general, pathologists with urologic subspeciality training show higher rates of interobserver agreement than general pathologists, and reviews by experts lead to more accurate risk stratification than reviews by less experienced pathologists."
In recent years, Google has been actively developing AI algorithms for a variety of healthcare applications, such as detecting breast cancer on mammograms, assessing cancer risk on computed tomography (CT) lung cancer screening exams, finding diabetic retinopathy on retinal scans, and predicting medical events through analysis of electronic medical record software.
Company researchers reported results for their deep-learning system in analyzing pathology images and predicting survival of patients with 10 cancer types in PLOS One in June.
False positives trade-off
In the latest study, subspecialists made their assessments using three histological sections plus an immunohistochemical stained section for each specimen. A majority opinion was determined. Performance was compared to the deep-learning system and evaluations from a panel of board-certified general pathologists. General pathologists and the algorithm worked without immunohistochemical stained sections, simulating routine workflow.
In addition to Gleason grading, the study evaluated performance of subspecialists, general pathologists, and the deep-learning system for differentiating specimens with and without cancer. A total of 752 specimens were assessed for this part of the study. And in this scenario, the performance of the general pathologists and the deep-learning system were on par; that is, in agreement with the subspecialist findings in 94.3% and 94.7% of cases, respectively. Compared with general pathologists, the deep-learning system caught more cancers, but also flagged more false positives.
"This trade-off suggests that the [deep-learning system] could help alert pathologists to tumors that may otherwise be missed, while relying on pathologist judgment to overrule false-positive categorizations on small tissue regions," the authors advised.
Overall, the results suggest that an automated system could bring performance closer to the level of experts and boost the value of prostate biopsies, in the authors' view.
"Future research is necessary to evaluate the potential utility of using the [deep-learning system] as a decision support tool in clinical workflows and to improve the quality of prostate cancer grading for therapy decisions," Mermel et al concluded.