Scientists have designed a nanopore-based sensor that can detect the presence and infectiousness of a virus in a sample within minutes, according to a new study published in Science Advances on September 22. The new method successfully detected SARS-CoV-2, positioning it as a speedier, less fussy alternative to other tests.
The COVID-19 pandemic has highlighted the need for rapid, sensitive tests of viral pathogens. Current COVID-19 tests are slow and do not differentiate between noninfectious and infectious viruses. For example, SARS-CoV-2 viral RNA has been detected in patients more than one month after the onset of illness even though the virus cannot be detected by culture after week three.
In the study, researchers designed a DNA aptamer-nanopore system (so named because it incorporates specially designed short single-stranded RNA or DNA oligonucleotides called aptamers into nanopore sensing) that is capable of detecting virus particles at extremely low levels in complex samples in as little as 30 minutes to two hours. Additionally, the samples do not need pretreatment to minimize false-negative and false-positive results.
Compared to the new sensor, quantitative polymerase chain reaction (PCR) tests require more time and expensive lab equipment to process the results, and they don't distinguish infectious samples (or identify contagious patients). The other tool microbiologists use is the plaque assay, which measures infectious viral particles but takes days to render results.
"With the virus that causes COVID-19, it has been shown that the level of viral RNA has minimal correlation with the virus' infectivity. In the early stage when a person is infected, the viral RNA is low and difficult to detect, but the person is highly contagious," Yi Lu, PhD, a professor emeritus of chemistry at the University of Illinois Urbana-Champaign, said in a statement.
When a person recovers from COVID-19, the viral RNA can be high even though the person is not infectious. A similar patten emerges -- but much later -- with antigen tests, Lu noted.
"Viral RNA and antigen tests are both poor in informing whether a virus is infectious or not," Lu said. "It may result in delayed treatment or quarantine, or premature release of those who may still be contagious."
DNA aptamer-nanopore sensors
To create the sensor, the scientists selected highly specific DNA aptamers from a DNA library to bind to intact infectious viruses while excluding noninfectious viruses. They focused specifically on detecting two different types of viruses: SARS-CoV-2 and human adenovirus (HAdV), an emerging waterborne viral pathogen.
Aptamers are well suited to the task because they bind target molecules with an affinity and selectivity that rivals antibodies, yet they are less expensive and easier to work with. The scientists obtained DNA aptamers for SARS-CoV-2 and HAdV synthetically in a test tube using a technique called systematic evolution of ligands by exponential enrichment (SELEX). The technique differentiates infectious and noninfectious viruses depending on the composition of the viruses' surface.
With the SARS-CoV-2 virus, the SELEX technique yielded a highly specific aptamer, SARS2-AR10, that was able to bind to active SARS-CoV-2 while ignoring harmless versions of the virus that had been inactivated by UV light and other coronaviruses, such as the 229E coronavirus.
With the necessary aptamers in hand, the scientists then incorporated them into a solid-state nanopore that allows strong confinement of the virus to enhance sensitivity down to 1 pfu/ml for HAdV and 1 × 104 copies/mL for SARS-CoV-2.
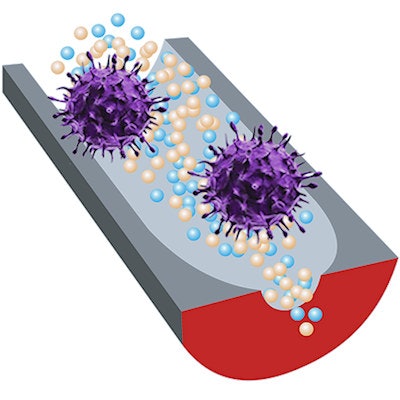
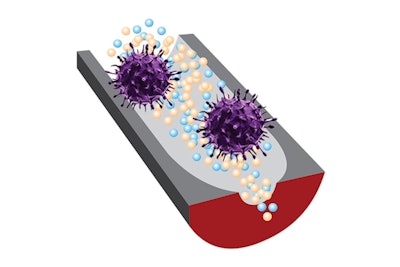 A new sensor can distinguish infectious viruses from noninfectious ones, thanks to selective DNA fragments and sensitive nanopore technology. Image courtesy of Ana Peinetti.
A new sensor can distinguish infectious viruses from noninfectious ones, thanks to selective DNA fragments and sensitive nanopore technology. Image courtesy of Ana Peinetti."Our sensor combines two key components: highly specific DNA molecules and highly sensitive nanopore technology," Ana Peinetti, PhD, the first author of the study and a researcher at the University of Buenos Aires, said. "We developed these highly specific DNA molecules, named aptamers, that not only recognize viruses but also can differentiate the infectivity status of the virus."
The sensing technique could be applied to other viruses, the researchers said, by tweaking SELEX to obtain DNA aptamers that target those viruses.
"The implementation of SELEX to obtain specific aptamers for infectious viruses ... makes it possible to quantify viruses in samples ranging from complex environmental samples to untreated biological fluids, which will allow a wide range of applications for rapid and selective detection of both current and emerging viral pathogens around the world," the authors wrote.
In the future, in addition to improving the sensitivity and selectivity of the sensor, the scientists hope to incorporate DNA aptamers into more user-friendly product formats, such as color-changing dipsticks or smartphone-compatible sensors.